Our team has a long-standing history with Clinical Outcome Prediction. From identifying the most valuable time for the task to multiple iterations to improve our models' performance, we published multiple peer-reviewed papers. The most noteworthy on the subject are:
Clinical Outcome Prediction from Admission Notes using Self-Supervised Knowledge Integration
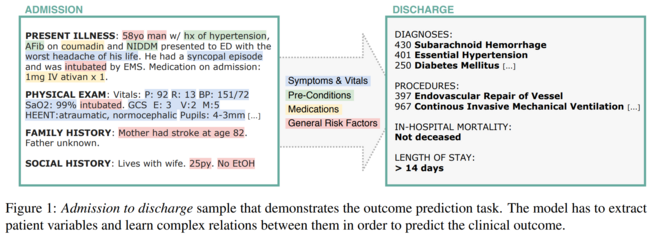
Abstract: Outcome prediction from clinical text can prevent doctors from overlooking possible risks and help hospitals to plan capacities. We simulate patients at admission time, when decision support can be especially valuable, and contribute a novel admission to discharge task with four common outcome prediction targets: Diagnoses at discharge, procedures performed, in-hospital mortality and length-of-stay prediction. The ideal system should infer outcomes based on symptoms, pre-conditions and risk factors of a patient. We evaluate the effectiveness of language models to handle this scenario and propose clinical outcome pre-training to integrate knowledge about patient outcomes from multiple public sources. We further present a simple method to incorporate ICD code hierarchy into the models. We show that our approach improves performance on the outcome tasks against several baselines. A detailed analysis reveals further strengths of the model, including transferability, but also weaknesses such as handling of vital values and inconsistencies in the underlying data.
Models on huggingface:
- Pre-Trained CORe Model
- Fine-Tuned CORe for Diagnosis Prediction
- Fine-Tuned CORe for Mortality Prediction
Betty van Aken, Jens-Michalis Papaioannou, Manuel Mayrdorfer, Klemens Budde, Felix Gers, Alexander Löser. 2021. Clinical Outcome Prediction from Admission Notes using Self-Supervised Knowledge Integration. In Proceedings of the 16th Conference of the European Chapter of the Association for Computational Linguistics: Main Volume, pages 881–893, Online. Association for Computational Linguistics.
This Patient Looks Like That Patient: Prototypical Networks for Interpretable Diagnosis Prediction from Clinical Text
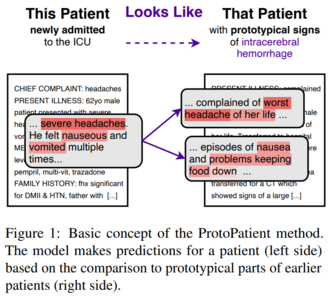
Abstract: The use of deep neural models for diagnosis prediction from clinical text has shown promising results. However, in clinical practice such models must not only be accurate, but provide doctors with interpretable and helpful results. We introduce ProtoPatient, a novel method based on prototypical networks and label-wise attention with both of these abilities. ProtoPatient makes predictions based on parts of the text that are similar to prototypical patients—providing justifications that doctors understand. We evaluate the model on two publicly available clinical datasets and show that it outperforms existing baselines. Quantitative and qualitative evaluations with medical doctors further demonstrate that the model provides valuable explanations for clinical decision support.
Betty van Aken, Jens-Michalis Papaioannou, Marcel Naik, Georgios Eleftheriadis, Wolfgang Nejdl, Felix Gers, and Alexander Loeser. 2022. This Patient Looks Like That Patient: Prototypical Networks for Interpretable Diagnosis Prediction from Clinical Text. In Proceedings of the 2nd Conference of the Asia-Pacific Chapter of the Association for Computational Linguistics and the 12th International Joint Conference on Natural Language Processing (Volume 1: Long Papers), pages 172–184, Online only. Association for Computational Linguistics.
Boosting Long-Tail Data Classification with Sparse Prototypical Networks
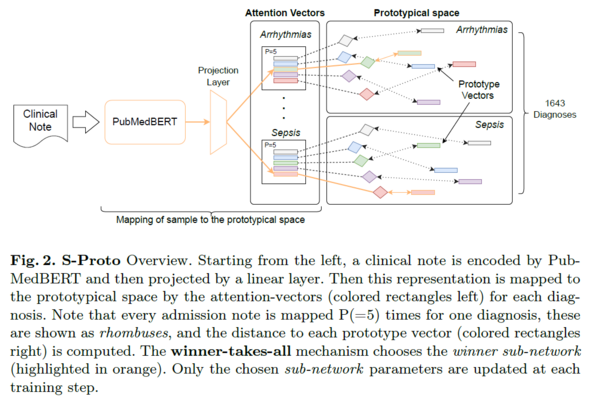
Abstract: Clinical Decision Support Systems (CDSS) have become ubiquitous in healthcare facilities, leveraging the increasing presence of Electronic Health Records (EHR). Predicting clinical outcomes from clinical text, such as identifying diagnoses based on the admission state of patients, is among the core tasks that a CDSS must address. The state-of-the-art for this task has been set by transformer encoder models, recently superseded by encoders enhanced with a prototypical network. This task remains a significant challenge due to the substantial imbalance of the outcome labels, which is characterized by a long-tailed distribution where the majority of diagnoses are under-represented. Motivated by recent biologically inspired findings in deep learning, we propose S-Proto, a novel, efficient, and sparse prototypical layer. Our method achieves state-of-theart performance in outcome diagnosis prediction, without compromising on the explainability characteristics of prototypical encoders. Quantitative results demonstrate that our approach is robust to the challenges presented by clinical notes, and transfers successfully to a second, unseen dataset. Qualitative evaluation with medical doctors shows that S-Proto is capable of disaggregating the representations of a disease that manifests differently in patient cohorts.
Alexei Figueroa, Jens-Michalis Papaioannou, Conor Fallon, Alexandra Bekiaridou, Keno Bressem, Stavros Zanos, Felix Gers, Wolfgang Nejdl, Alexander Löser. 2024. Boosting Long-Tail Data Classification with Sparse Prototypical Networks. Manuscript submitted for publication.
